In eukaryotes, the expression of protein-coding genes involves the integration of cellular signals, chromatin modification, and assembly of the transcription pre-initiation complex (PIC) that loads RNA polymerase II onto the core promoter. PIC formation is initiated by recruitment of the TATA-binding protein (TBP) to the promoter by protein complexes containing TBP-associated factors (TAFs) that interact with activators, promoter DNA, and/or chromatin. Throughout evolution, these TAFs partitioned and specialized into two distinct coactivator complexes in eukaryotes, the general transcription factor TFIID and the Spt-Ada-Gcn5 acetyltransferase (SAGA) complex (Fig. 1). Our previous studies on human TFIID (hTFIID) have elucidated its molecular function as a chaperone for TBP deposition and initiation of PIC assembly.
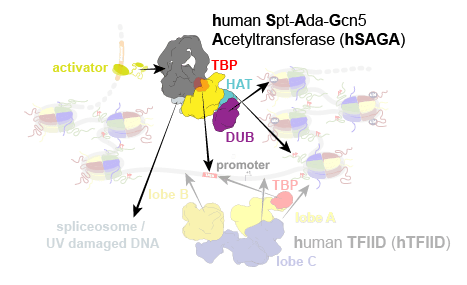
Studies of TFIID and SAGA in yeast highlight these TAF-containing complexes as global coactivators of transcription, but their unique or overlapping roles in human gene regulation are still not well understood. SAGA has been functionally implicated in a multitude of cellular pathways, from serving as a regulatory hub in transcription to its involvement in cell proliferation. With more than a megadalton in size, SAGA contains several distinct functional modules (Fig. 2):
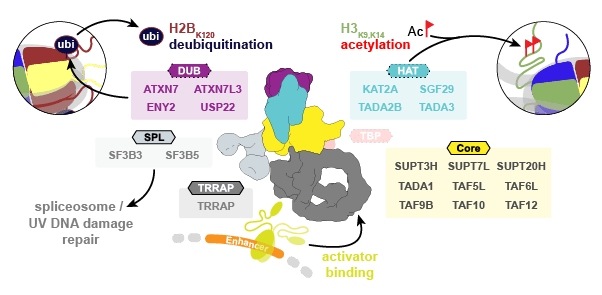
A core module that forms a structural scaffold of histone-fold (HF)-containing TAFs; a TRRAP (Transformation/Transcription domain Associated Protein) module that contains a phosphoinositide-3 kinase (PI3K)-related pseudo protein kinase (ΨPIKK) and binds activators such as c-Myc, E2F, and p53; a histone acetyltransferase (HAT) module that deposits H3K9ac and H3K14ac at promoters of active genes; and a deubiquitinase (DUB) module that removes H2B K120 ubiquitination from active gene bodies.
Initial structural studies on SAGA were limited to isolated regions or domain fragments. More recent studies on the complete yeast SAGA (ySAGA) complex have revealed its modular organization, and structural details for two of its modules. While vertebrate SAGA is highly conserved, the conservation with yeast drops dramatically and numerous domain insertions and deletions as well as gene duplication, have led to the subfunctionalization of human SAGA (hSAGA) subunits. Substantial evolutionary differences are further indicated by the lack of a metazoan homolog to one TBP-binding subunit in ySAGA (Spt8), and by the incorporation of the splicing factor subunits SF3B3 and SF3B5 within the metazoan complex, which gives rise to a fifth, splicing module (SPL) in metazoan SAGA (Fig.2). Lastly, while SAGA is not essential for viability in yeast, it is strictly required for metazoan development, and disruption of hSAGA can lead to broad pathogenic effects, including neurodegenerative disease and cancer. The difference in its essentiality, as well as the presence of specialized subunit paralogues and gained interaction partners, indicate that some aspects of assembly and function must be clearly different between the human and yeast SAGA complexes.
To study the architecture of hSAGA and to examine the possible functional implications of its conserved and distinct features, we determined the structure of hSAGA using negative stain and cryo-electron microscopy at 19 Å and 2.9 Å resolution, respectively (Video 1).
The 20 subunit hSAGA complex reveals a divergent architecture, that integrates the metazoan specific U2 splicing subunits and lacks the essential subunit for TBP binding in yeast. While the core module contains a distorted histone fold octamer like the one observed in yeast, the domain topology in its periphery is different, which causes global architectural differences and is key for the integration of the splicing module. The most significant difference is the attachment site of the TRRAP module, which is rotated around the core by almost 90° (Video 2). In contrast to ySAGA, where only two to three subunits contribute to the formation of a flexible core-Tra1 (yeast homolog of TRRAP) interface, all almost all core subunits (eight) contribute to interface formation in hSAGA. A transition between the yeast and human conformation is highly unlikely since this would require unfolding of essential structural elements.
A major contributor to the diverging architecture is the subunit SUPT20H that contributes to the architecture in the core module with its N-terminal domain and contains a long linker “the latch” that wraps around the FAT domain of the TRRAP module and connects a C-terminal domain (CTD) with structural homology to an SH2 domain. Both, the latch and CTD are unique for metazoan SAGA and the CTD is bound in a pocket below the FAT domain at the entrance towards a highly positively charged tunnel (Fig. 3).
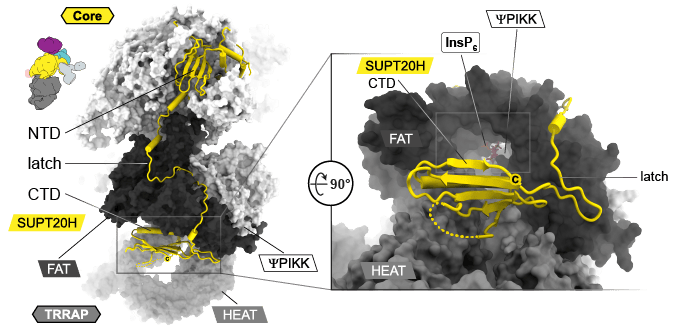
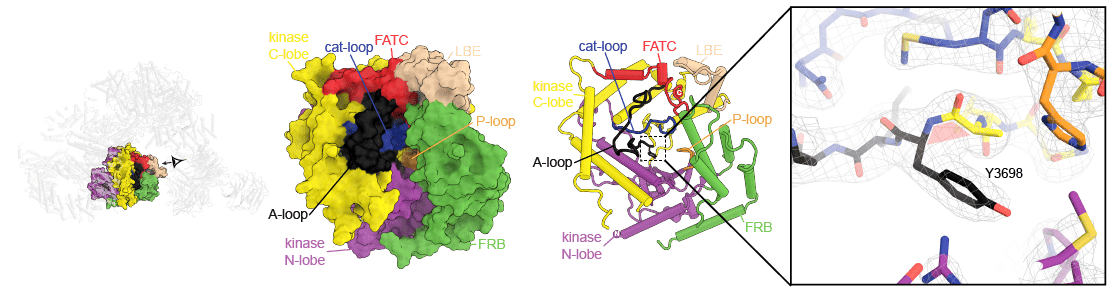
Bound in this tunnel is an inositol hexakisphosphate (InsP6), which is well-defined in the EM-map. The tunnel is situated in the back of the ΨPIKK. Due to the absence of the for catalysis crucial “DFG” motif in active kinases, the enzyme was described as inactive. In active kinases, the aspartate (D) of this motif would interact with the pyrophosphate group of ATP. However, the high resolution of our structure identified that the corresponding residue in the center of the active site (Y3698) adopts an unusual cis-peptide bond. Together with remarkably high sequence conservation of the ΨPIKK domain (~65%) in metazoan (25% in yeast), this high energy state in the center of an active site might indicate a so far undiscovered function apart from phosphorylating substrates (see https://doi.org/10.1016/j.tcb.2014.03.008) (Fig. 4).
Although TBP binding has not been observed for metazoan SAGA, it is well described for ySAGA, where Spt8 is essential for TBP binding, which does not exist in metazoan. Interestingly, our structure also revealed the location of a number of disease mutations and several activator binding sites. One of them is c-Myc, which is known to bind TBP and hSAGA via non-overlapping regions. This raises the question of whether activators can contribute to TBP recruitment to metazoan SAGA.
In yeast, a possible mechanism (https://doi.org/10.1038/s41586-020-1944-2) for TBP loading onto a promoter was discussed, but the structure indicated that although DNA would fit in the gap between the ySAGA core and TBP its path would clash with the location of the Tra1 module. However, in hSAGA this path is fully accessible (Fig. 5).

Our cryo-EM structure of hSAGA allows mapping of human disease mutations and paves the way for structure-guided drug design of this important therapeutic target for human developmental diseases and cancer.

