Plants and animals respond to pathogen invasion via intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) that directly interact with pathogen proteins or indirectly detect pathogen derived alteration in the host proteome. Upon recognition of pathogen invasion, NLRs trigger an immune response that resolves in a variety of ways depending on the type of NLR being activated. The overall architecture of NLRs is highly conserved, consisting of an C-terminal LRR platform that determines substrate specificity and a central nucleotide-binding oligomerization domain. The N-terminal domain varies between NLRs and determines the mechanism utilized by the host to activate the immune response. Thus, NLRs in plants have been classified according to their N-terminal domain into Toll/interleukin 1 receptor NLRs (TNLs), coiled-coil NLRs (CNLs) and RPW8-like coiled-coil NLRs (RNLs). Pathogen detection and oligomerization of the NLR activates these N-terminal domains by bringing them in close-contact. In all 3 cases, association of the N-terminal domain leads to localized cell death and expression of disease resistance. The TIR domains of TNLs have been shown to possess oligomerization-dependent NADase activity that is required for promoting cell death, yet it is not understood how the interactions between TIR domains renders them catalytically active.
The structure of the ROQ1-XopQ complex, an immune receptor bound to its pathogen substrate, was used as a model to study the mechanism of direct binding, oligomerization, and TIR domain activation of TNLs. ROQ1 has been shown to physically interact with the Xanthomonas effector XopQ, causing it to oligomerize and trigger a TIR-dependent hypersensitive cell death response. We co-expressed, extracted and purified the assembled ROQ1-XopQ complex from ROQ1’s native host, Nicotiana benthamiana, and solved its structure by cryo-EM to 3.8 Å resolution. The interactions described in our structure were further confirmed by in vivo mutational analysis.
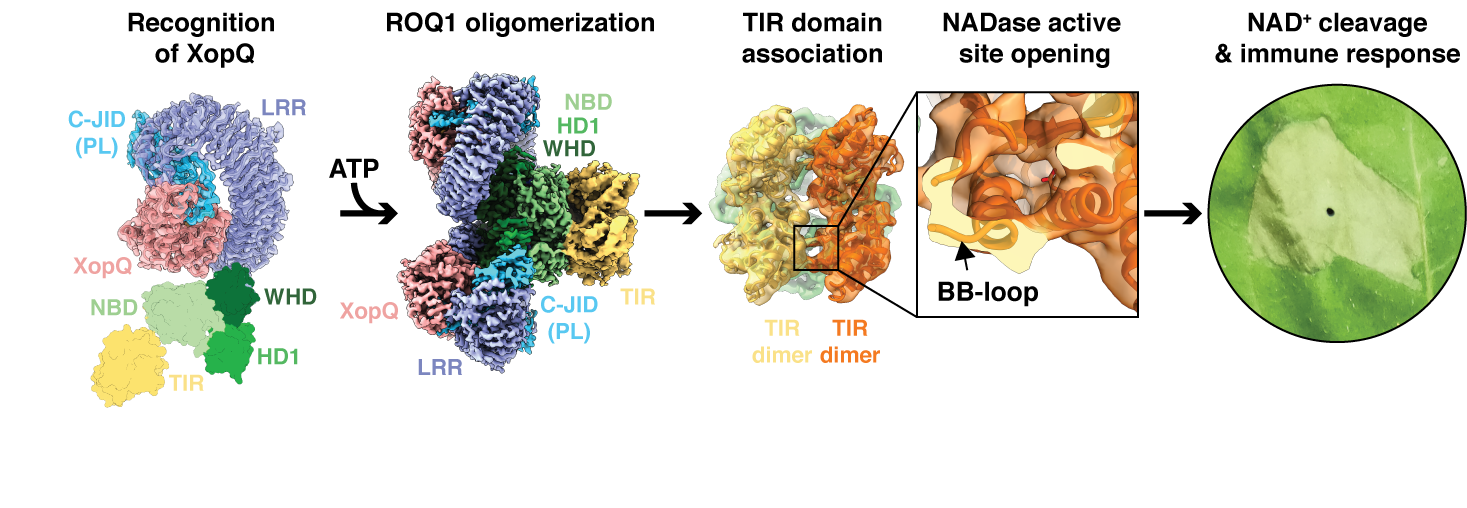
Our structure reveals that ROQ1 forms a tetrameric resistosome upon recognizing XopQ.
The LRR and a post-LRR domain, termed C-JID (C-terminal Jelly-roll/Ig-like Domain), form a horseshoe shaped scaffold that curls around the pathogen effector, thereby recognizing multiple regions of the substrate. Binding of the ROQ1 LRR to XopQ occurs via surface exposed residues that make up the scaffold of the domain, as well as an elongated loop between two leucine-rich repeats that forms a small amphipathic α-helix at the site of interaction. The mode of substrate recognition by the C-JID is reminiscent of that used by immunoglobulins to bind their antigen. Similarly to the complementary-determining regions of antibodies, interconnecting loops emerging from the C-JID β-sandwich structure make substrate-specific contacts with XopQ. In particular, an extended loop of the C-JID dives into the active site cleft of XopQ and interacts with conserved residues required for nucleoside binding, suggesting that ROQ1 not only recognizes its substrate but also inhibits its ligand-binding function.
The NB-ARC domain (NBD, HD1, WHD) of ROQ1 responsible for oligomerization is found in an ATP-bound state. Individual protomers intercalate in a similar fashion as found in other NLR structures, promoting association between the N-terminal TIR domains. The TIR domains bind to each other via two distinct interfaces (termed AE and BE) causing them to form a dimer of dimers. BE-interface contacts cause a conformational rearrangement in a loop, termed BB-loop, at the periphery of the TIR domain active site that exposes the putative catalytic glutamate, suggested to cleave NAD+. These results provide a rationale for the previously determined oligomerization-dependence of TIR domain NADase activity.
We propose a step-by-step mechanism for ROQ1 immune signaling based on our structure of the activated complex and previous biochemical studies. The LRR and C-JID of ROQ1 recognize the pathogen effector via direct contacts with its surface and active site residues. Detection of the substrate releases autoinhibitory contacts between the NB-ARC domain and the LRR allowing the NB-ARC domain to transition to an ATP-bound oligomerization prone state. Complex assembly brings the TIR domains in close contact leading to opening of the NADase active site in an interface-dependent manner. Cleavage of NAD+ by the TIR domain results in the release of ADP-ribose, a signaling molecule that triggers cytosolic calcium influx, a widely utilized chemical cue in response to various biotic and abiotic stresses, leading to downstream activation of localized cell death and disease resistance.