The proper segregation of chromosomes during cell division is fundamental for all living organisms to maintain genome integrity. Kinetochores are large protein assemblies that connect chromosomes to spindle microtubules (MTs) that drive chromosome separation during mitosis. Lengthening and shortening of microtubules exert forces on kinetochores that are coupled to chromosome movement. In humans, kinetochores consist of more than eighty proteins, forming two structural layers: a “inner kinetochore” region called CCAN (Constitutive Centromere Associated Network) that interacts with specialized chromosome regions called centromeres, and an “outer kinetochore” region that attaches to microtubules, called the KMN network (KNL1, Mis12, and Ndc80 complexes). During chromosome segregation, kinetochores interacting with centromeres switch from lateral interactions with microtubules to “end-on” interactions, where the plus end of the microtubule is “embedded” in the kinetochore. This “end-on” attachment then couples de-polymerization of microtubule ends to chromosome movement.
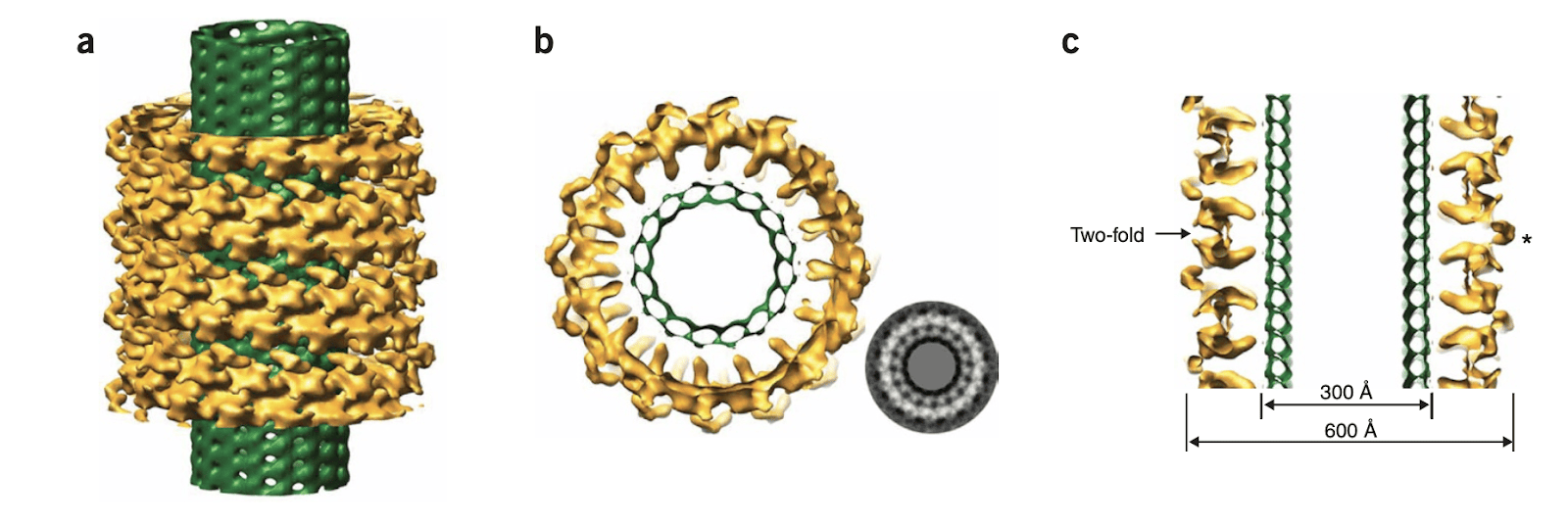
Understanding how kinetochores maintain stable attachment to the microtubule ends, and how these interactions are modified to support the addition and removal of tubulin molecules has been a long-standing question in the field. We have been studying the yeast Dam1 kinetochore complex for more than a decade now. Our initial studies, in collaboration with the Drubin and Barnes labs (UC Berkeley), showed that this complex assembles into rings around microtubules and that the rings move processively with microtubule ends, coupling microtubule depolymerization to directional movement. We used EM and image reconstruction to produce some of the first structures of the Dam1 complex on its own and bound to microtubules (Westermann et al., Mol. Cell 2005, Westermann et al., Nature 2006, Wang et al., NSMB 2007).
We further characterized the yeast Ndc80 kinetochore complex, the microtubule interacting component of the KMN that interacts with the Dam1 complex. We visualized the full-length yeast Ndc80 complex and found a dramatic kink within the 560-Å complex localized to a conserved break in the coiled-coil and proposed its important in kinetochore geometry and likely as part of a tension sensing mechanism (Wang et al., JMB 2008). Using a minimal bonsai construct of the huma Ndc80 complex, we obtained a subnanometer structure of Ndc80 bound to the microtubule (Alushin et al. Nature 2010). The complex binds with a monomeric tubulin repeat, both at intra- and inter-dimer interfaces, using a minimal “toe-print” that reads highly conserved sequences in tubulin. We also dissected the role of the unstructured N-terminus of Ndc80, a major site of Aurora B phosphorylation, leading to a model of how Ndc80’s interaction with MT ends is tuned by the phosphorylation state of its tail. In the process, we have obtained the first structure of the C-terminal tail of tubulin, as it engages the Ndc80 complex in an adjacent protofilament (Alushin et al. NSMB 2012).
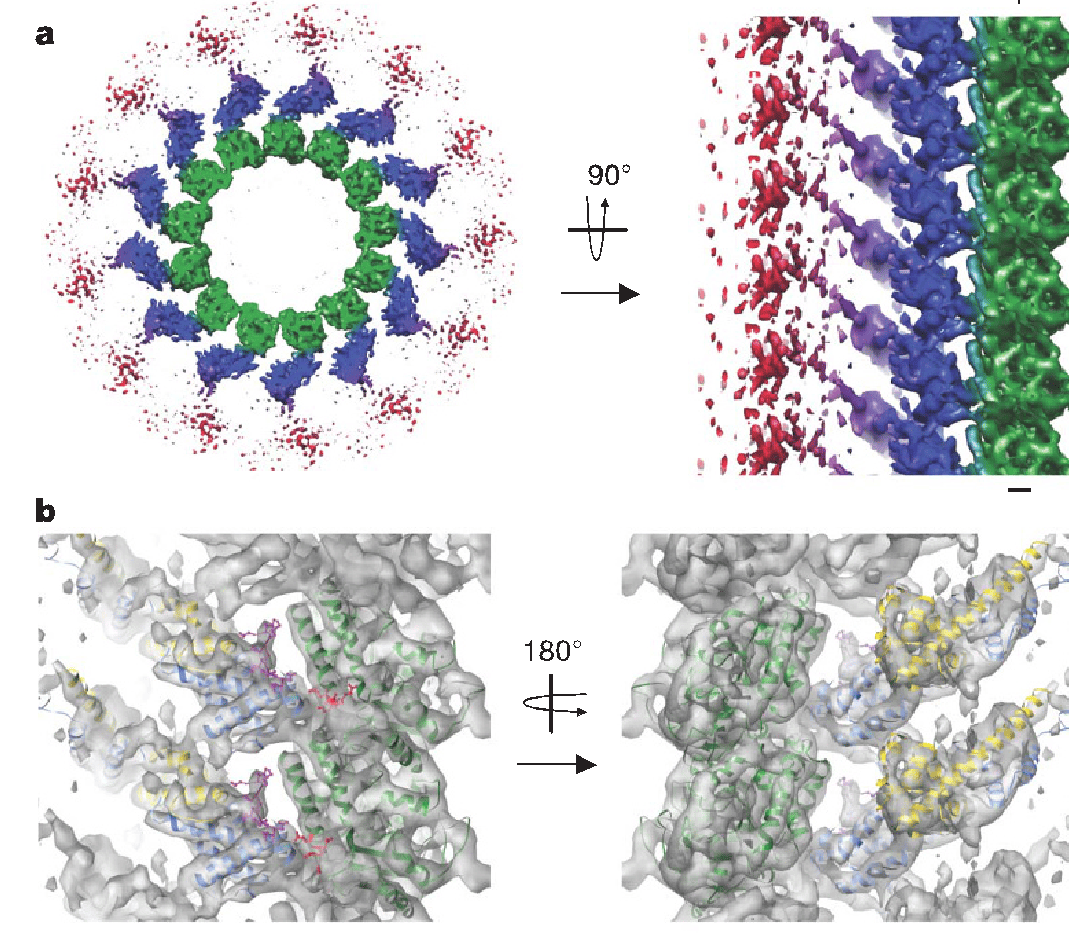
We have now extended our studies to include the mammalian orthologs of the Dam1/Ndc80 complex and are combining techniques in cryo-EM and cryo-ET to understand kinetochore-MT connections in human cells. Together we hope to better understand the conserved mechanisms for faithful chromosome attachment during mitosis that is essential for cellular survival and genome integrity.
