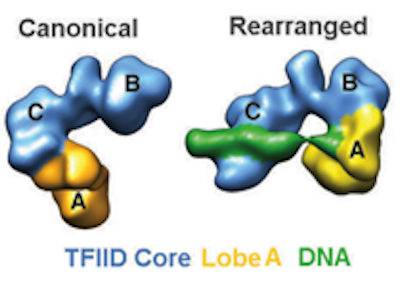
The accurate initiation of transcription requires the assembly of a pre-initiation complex (PIC) that include TFIID, TFIIA, TFIIB, TFIIE, TFIIF, TFIIH and RNA pol II. Regulation is achieved by gene specific activators and repressors, cofactor complexes that mediate the interaction of the general machinery with sequence-specific activators, and protein complexes involved in the modification or remodeling of chromatin. The Nogales lab is interested in characterizing the structure of these different components and how they interact to regulate transcription. A main effort has been to define the structure of the human transcription factor IID (TFIID). Binding of this general factor to the core promoter is the first step in the assembly of the whole transcriptional machinery. In collaboration with Robert Tjian (UC Berkeley) we obtained the first 3-D model of TFIID and showed the existence of significant flexibility within the complex, which we proposed could play a distinct role in directing the formation of an active PIC. We also characterized a cell type-specific TFIID complex containing TAF4b and studied the interaction of TFIID with differential activators. In exciting and recent work in collaboration with James Kadonaga (UCSD), we has shown that TFIID coexists in two predominant states differing dramatically in the location of lobe A (containing TBP and TFIIA) with respect to a more stable BC core. A novel conformation of TFIID, the rearranged state, interacts with promoter DNA in a TFIIA-dependent manner. We found that the downstream region of the SCP is bound by lobe C, while the upstream DNA sequence is bound within lobe A. This has lead us to propose that the dynamic conformational landscape of TFIID may have regulatory consequences by providing specific structural targets that can be recognized by transcriptional activators and repressors. Testing this idea is a major, on going effort.
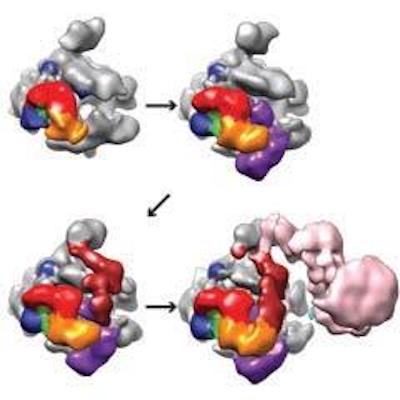
Recently, the Nogales lab has developed an in vitro reconstitution system to describe thestepwise assembly of the human PIC. This study allowed us to describe how TFIIF stabilizesthe core promoter DNA along the surface of RNAPII, and how TFIIE addition results in thetopological trapping of the DNA on the RNAPII cleft. TFIIE positions TFIIH so that the activeATPase in transcription initiation, XPB, is down stream of the transcription start site. We alsoused an artificial DNA template that served as a mimic of that generated naturally by thehelicase action of TFIIH. The apparent movementof downstream DNA in this structure, togetherwith the positioning of XPB, suggests how XPB would act as a DNA translocase whose activitywould push against the stably bound upstream DNA at the TATA box to induce negativesupercoiling at the transcriptionstart site.
See also:
Liu, W-L., Coleman, R.A., Grob, P., Geles, K.G., King, D.S., Ramey, V.H., Nogales, E. and Tjian, R. (2008) Structural changes in TAF4b-TFIID correlated with promoter selectivity. Mol Cell., 29, 81-91.
Liu, W-L., Coleman, R.A., Yang, J-L., Grob, P., Ma, E., Dailey, G., Nogales, E., and Tjian, R. (2009) Structures of three distinct activator-TFIID complexes. Genes Dev., 23(13), 1510-1521.
Cianfrocco, M.A., Kassevitis, G.A., Grob, P, Fang, J., Juven-Gershon, T., Kadonaga, J.T. and Nogales, E. (2013) Human TFIID binds core promoter DNA in a reorganized structural state. Cell, 152(1-2)120-131
He, Y., Fang, J., Taatjes, D.J., and Nogales, E. (2013) Structural visualization of key steps in human transcription initiation. Nature,495(7442)481-6
Michael A. Cianfrocco and Eva Nogales (2013) Regulatory interplay between TFIID’s conformational transitions and its modular interaction with core promoter DNA Transcription Transcription., 2013 Jun 21, Epub ahead of print
