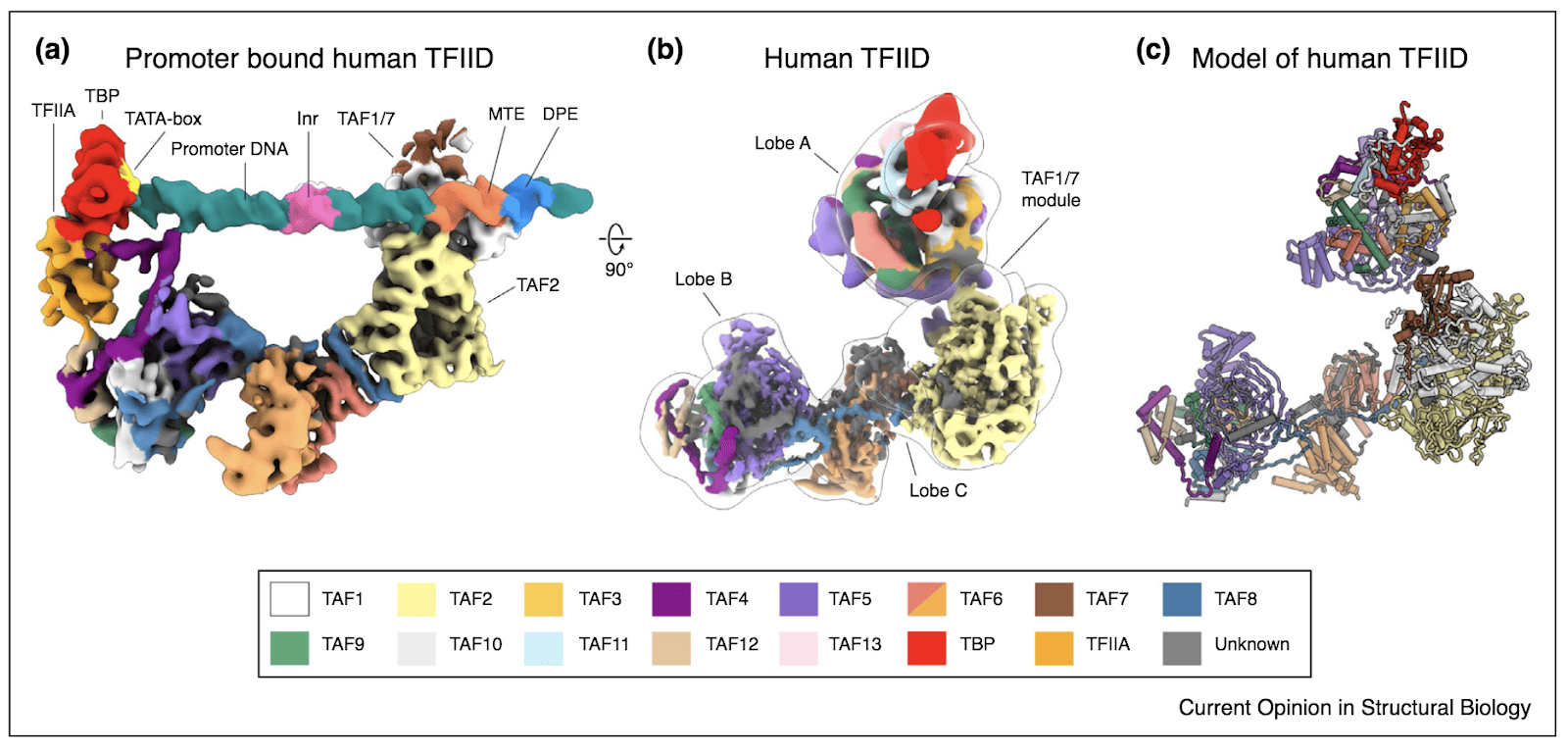
Eukaryotic transcription of protein-coding genes requires the assembly of a Pre-Initiation Complex (PIC) at the core promoter. This large macromolecular complex includes RNA Polymerase II (RNA Pol II) and several general transcription factors. The factor that nucleates the assembly of the PIC is TFIID, a multi-subunit complex containing the TATA-binding protein (TBP) and several TBP-associated factors (TAFs). For a long time, structural studies on this complex have been hindered by three main challenges: the scarcity of the sample, the poor stability of the complex, and its intrinsic flexibility. Overcoming these technical challenges, our lab presented the first complete model of human TFIID and provided structural insight on the mechanism by which TBP is loaded to the promoter DNA.
The architecture of TFIID is organized in a tri-lobular fashion, with a relatively rigid core formed by lobes B and C, and a highly flexible lobe A. Lobe C contains DNA-binding elements that can engage conserved downstream regulatory motifs in the promoter, supporting the role of the BC core as a molecular ruler that enables accurate TBP deposition on the DNA. Interestingly, the dynamic nature of lobe A in TFIID plays a critical role in sampling the conformational space of the system and provides a mechanistic explanation for TATA-box scanning, recognition and engagement by TBP.
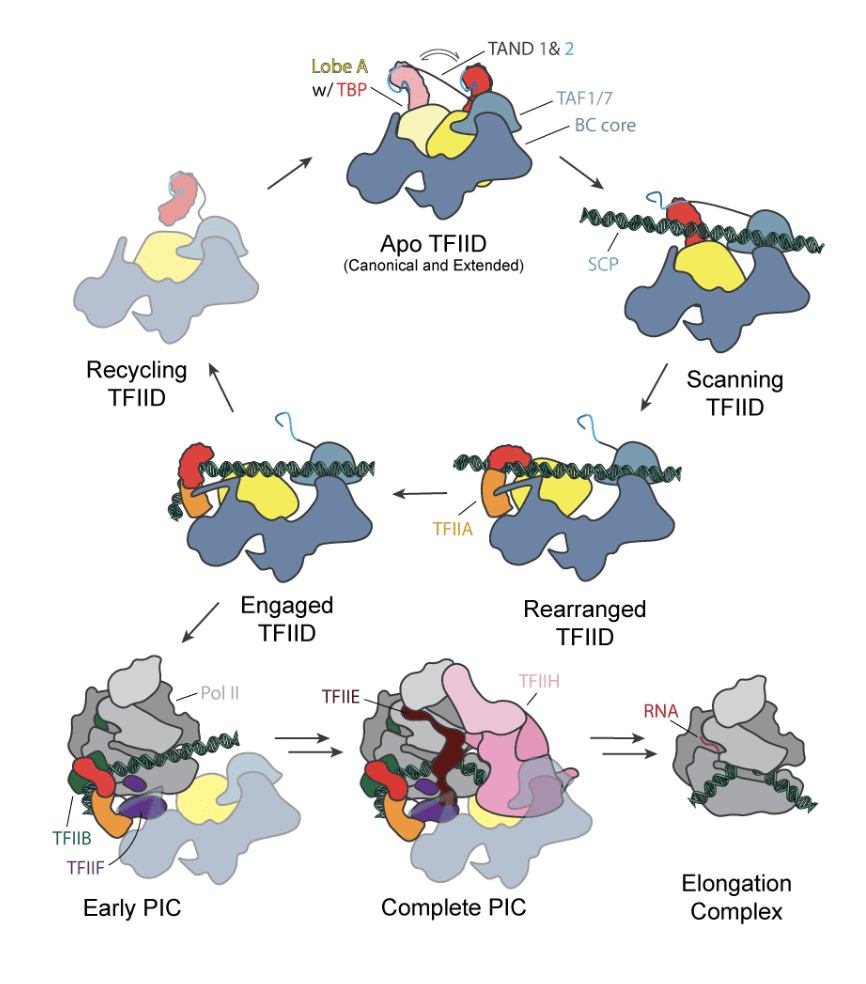
In our study, we were able to distinguish between two major states for apo-TFIID (termed the canonical and extended states) through computational sorting of the cryo-EM data. In the context of core promoter DNA and TFIIA –the second general transcription factor to be recruited in the stepwise assembly of the PIC–, we observed three additional states (termed the scanning, rearranged, and engaged states). Lobe A, which migrates 150 Å from its position near lobe C in the canonical state to near lobe B in the extended state, carries TBP in a repressed state that is only released in the context of promoter binding. Identification of distinct TFIID states allowed us to generate a mechanistic model for TFIID promoter binding (see figure below). We proposed that TFIID first binds the downstream core promoter elements through TAF1 and TAF2. This binding and the flexible attachment of lobe A help position the upstream DNA in proximity to TBP. TBP then scans for a TATA box or its sequence variants. Engagement of upstream core promoter sequences by TBP is facilitated by TFIIA interacting with TAF4 and TAF12 within lobe B. When TBP finally binds the promoter, it releases from lobe A, opening the binding site for TFIIB, which can then recruit Pol II.
- B., P., A. et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 (2018).
- K., L., R. et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–9 (2016).
- Nogales, E., Louder, R. K. & He, Y. Cryo-EM in the study of challenging systems: the human transcription pre-initiation complex. Curr Opin Struc Biol 40, 120–127 (2016).
