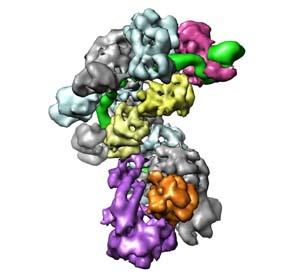
In bacteria and archaea, the clustered regularly interspaced short palindromic repeats (CRISPR)–associated (Cas) targeting complexes use CRISPR RNA (crRNA) guides to bind and destroy complementary nucleic acid targets. In the Type I-E CRISPR-Cas system in E. coli, these CRISPR transcripts are processed into short RNAs that are incorporated into a large ribonucleoprotein surveillance complex called Cascade. This complex specifically targets and binds foreign nucleic acids in a sequence-specific manner, leading to degradation of the targeted substrate. In collaboration with the Jennifer Doudna lab, we used cryo-electron microscopy to determine the structure of Cascade before and after binding to a ssRNA target substrate. These structures were solved to subnanometer resolution, and revealed that the CRISPR RNA (crRNA) is displayed along a helical arrangement of six CasC subunits. It was previously established that the Cascade complex engages invading nucleic acids through high-affinity base pairing interactions located near the 5’ end of the crRNA, but we determined in our studies that this binding triggers a concerted conformational change of the seahorse-shaped multisubunit complex. Together, these structures explain the molecular mechanism for target recognition and reveal a structural reorganization that likely serves as a signal for subsequent destruction of the invading DNA.
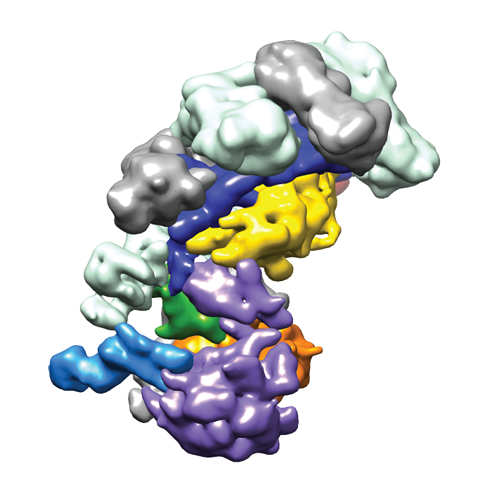
In vivo, Cascade targets double-stranded DNA (dsDNA) and recruits Cas3, a nuclease-helicase that catalyzes unwinding and cleavage of foreign dsDNA bearing a sequence matching that of the crRNA. Recently, we used single-particle electron microscopy reconstructions of dsDNA-bound Cascade with and without Cas3 to reveal that Cascade positions the PAM-proximal end of the DNA duplex at the CasA subunit and near the site of Cas3 association. The finding that the DNA target and Cas3 colocalize with CasA implicates this subunit in a key target-validation step during DNA interference. We show biochemically that base pairing of the PAM region is unnecessary for target binding but critical for Cas3-mediated degradation. In addition, the L1 loop of CasA, previously implicated in PAM recognition, is essential for Cas3 activation following target binding by Cascade.
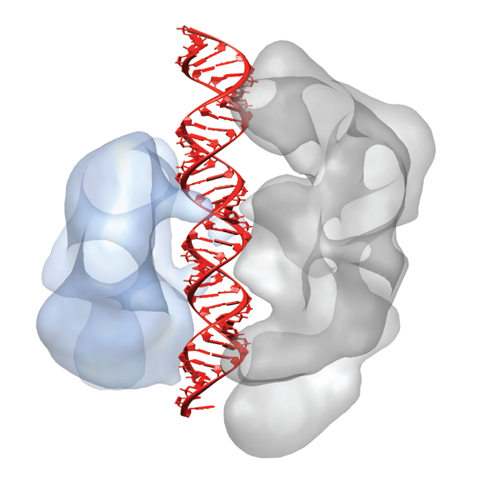
In Type II CRISPR-Cas systems, a single enzyme Cas9 uses a dual-RNA guide to perform sequence-specific cleavage of target DNA. Cas9:RNA also functions as a robust tool for both genome engineering and site-specific gene regulation in various organisms and cell lines. Recently, we showed that, in addition to providing base-pair recognition of target DNA sequences, the guide RNA triggers a conformational change in Cas9 that activates the complex for DNA surveillance. Single-particle electron microscopy reconstructions and site-specific labeling revealed that Cas9 consists of two structural lobes that undergo guide RNA-induced reorientation to create a central channel where DNA substrates are bound. The discovery that extensive structural rearrangements occur before target binding implicates RNA loading as a key regulator of enzyme function.
We are currently working towards a more complete structural understanding of CRISPR-Cas systems in collaboration with the Doudna lab.
